Abstract
Background: The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor critical for cellular metabolism, stem cell differentiation and immune control. HLA-E is a non-classical HLA molecule that is critical for NK cell activation. HLA-E is expressed on some AML blasts but genomic characterization is limited. IK-364, a tool compound generated by Ikena Oncology, is an AHR inhibitor in preclinical investigation. We set out to identify the genomic signatures associated with activation of the AHR pathway in AML samples utilizing the Beat AML dataset.
Methods: Whole bone marrow or peripheral blood samples were procured from AML patients under IRB-approved protocol (eIRB# 4422). Utilizing the Beat AML database, all samples were examined based on RNA AHR expression levels (RPKM) and correlated with genomic expression. High vs. low AHR expression was based on 90 th and 10 th percentiles. Pathway enrichment from upregulated genes was identified and compared between high vs. low AHR expression. Single cell RNA sequencing by 10x genomics Chromium system by the Massively Parallel Sequencing Shared Resource at OHSU was performed with previously frozen FLT3 positive AML samples (4 high AHR and 4 low AHR). Immunophenotyping was conducted by flow cytometry. The AHR antagonist IK-364 was utilized to study the in vitro effects of AHR modulation. Cytotoxicity was measured by flow cytometric detection of caspase-3/7 and SytoxAAD activation.
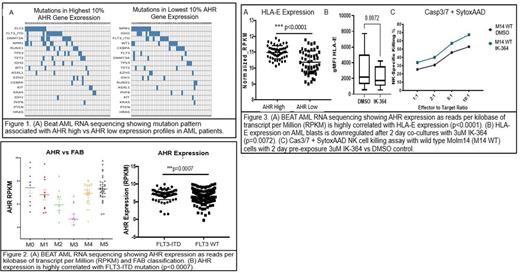
Results: The highest and lowest 10% expression levels of AHR in all samples in Beat AML were evaluated for mutation profiles. Ranking the top 18 mutations based on frequency we found an enrichment of FLT3 mutations in the high AHR expression group compared to the lower expression group (Figure 1A). ELN risk did not correlate with AHR expression however FAB classification M0/M1 and M4/M5 monocytic subtypes had higher AHR expression than M3 FAB subtype (Figure 2A). AHR gene expression was highly correlated with FLT3-ITD mutations (p=0.0007) (Figure 2B) and HLA-E expression (p=<0.0001) (Figure 3A). High AHR and high HLA-E joint expression in AML was associated with pathway upregulation in translocation of ZAP70 to immunological synapse, upregulation of HLA class II antigen presentation, phosphorylation of CD3 and TCR zeta chain and PD-1 signaling (p-value-1.11E-16), which are critical for innate and adaptive immune responses. AHR low and HLA-E low AML samples showed pathway enrichment for down-regulated genes including Hedgehog on state and neurexins and neuroligins (p value-0.001 and 7.61E-04 respectively). Genes upregulated in AHR high samples included CD14, CD86, HLA-DR, LILRB4, MAFB which are associated with monocytic phenotype. AHR high FLT3+ AML samples had higher protein expression of HLA-E on CD45+ blasts (p=0.03) compared to AHR low FLT3+ AML samples. Pretreatment of FLT3+ AML primary patient samples with IK-364 for 48 hours downregulates HLA-E on blasts (p=0.007) and, in a FLT3 positive AML cell line, Molm14, enhanced susceptibility to NK-cell-mediated killing (Figure 3B, 3C).
Discussion: Overall, we found that high AHR gene expression in AML correlates with FLT3-ITD expression and HLA-E as well as monocytic subtype. Pathway enrichment highlight the important role of AHR in immune regulation. Furthermore, AHR antagonism by IK-364, downregulates HLA-E on AML blasts and augments NK cell mediated killing of a mutant FLT3-ITD positive AML cell line. Our results highlight a continued interest in targeting the AHR pathway to augment immune-based therapies in AML.uture studies are underway to determine the mechanism behind this critical immune regulation.
Saultz: IKENA: Research Funding. Lind: IKENA: Research Funding. Tyner: Seattle Genetics: Research Funding; Constellation: Research Funding; Agios: Research Funding; Incyte: Research Funding; Petra: Research Funding; Gilead: Research Funding; Takeda: Research Funding; Janssen: Research Funding; Genentech: Research Funding; Array: Research Funding; Schrodinger: Research Funding; Astrazeneca: Research Funding. Kosaka: IKENA: Research Funding. McGovern: IKENA: Current Employment. Wang: IKENA: Current Employment. Sanchez-Martin: IKENA: Current Employment.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal